1 Trillion Dock
More than 70 billion molecules will be simulated on the 15 active interaction sites of the virus for a total of more than a thousand billion interactions evaluated in just 60 hours. This will be possible thanks to the simultaneous availability of the computing power (81 petaflops: millions of billions of operations per second) of Eni's HPC5, the most powerful industrial supercomputer in the world, of CINECA's Marconi100, and the virtual screening software accelerated by the Politecnico di Milano and Cineca, and the Exscalate molecular library from Dompé.
(*)The counter is updated considering the estimated processing speed obtained on the two supercomputers.
The helicase nsp13 catalyzes the unwinding of oligonucleotides duplex into single strands. Given its vital role in virus replication, this has been pointed as a promising pharmacological target. For this reason, a homology model was built using the crystal structure of MERS-CoV nsp13 (PDB Id: 5wwp) as the template. The orthosteric site is highly conserved and a good agreement between the binding site residues of the crystal structure and the SARS-CoV-2 homology model was found. A putative allosteric site for nsp13 was also derived from the NS3 helicase of HCV (PDB Id: 4B75). A crystal structure of the SARS-CoV-2 Helicase was deposited very recently (PDB Id: 6ZSL). HM 3D of structure of the SARS-CoV-2 Helicase NSP13 https://modelarchive.org/doi/10.5452/ma-epkbe
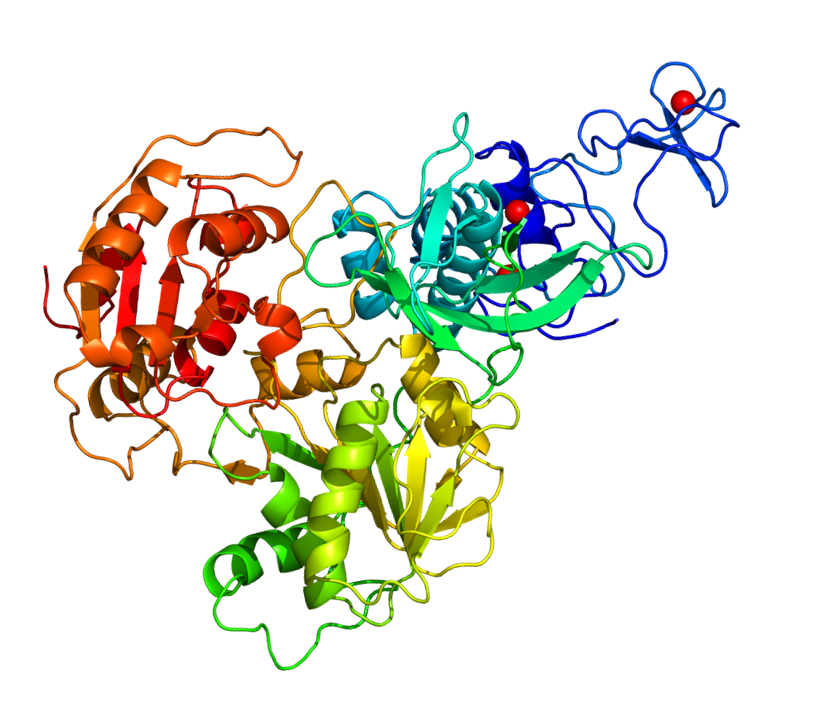
The helicase nsp13 catalyzes the unwinding of oligonucleotides duplex into single strands. Given its vital role in virus replication, this has been pointed as a promising pharmacological target. For this reason, a homology model was built using the crystal structure of MERS-CoV nsp13 (PDB Id: 5wwp) as the template. The orthosteric site is highly conserved and a good agreement between the binding site residues of the crystal structure and the SARS-CoV-2 homology model was found. A putative allosteric site for nsp13 was also derived from the NS3 helicase of HCV (PDB Id: 4B75). A crystal structure of the SARS-CoV-2 Helicase was deposited very recently (PDB Id: 6ZSL).HM 3D of structure of the SARS-CoV-2 Helicase NSP13 https://modelarchive.org/doi/10.5452/ma-epkbe
